Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Advertisement
Carousel with three slides shown at a time. Use the Previous and Next buttons to navigate three slides at a time, or the slide dot buttons at the end to jump three slides at a time.
02 November 2022
Wolfgang Lugmayr, Vadim Kotov, … Thomas C. Marlovits
14 June 2019
Jakob Runge, Sebastian Bathiany, … Jakob Zscheischler
24 October 2022
V. E. Ambruş, L. Bazzanini, … R. Tripiccione
14 November 2022
Robert Crystal-Ornelas, Charuleka Varadharajan, … Deborah A. Agarwal
14 October 2022
Trevor Barnes, Abhishek Shivakumar, … Taco Niet
29 October 2021
Gilles Maussion, Rhalena A. Thomas, … Thomas M. Durcan
11 August 2021
Maria Jardim Beira & Pedro José Sebastião
01 October 2021
Enrique Blanco, Mar González-Ramírez & Luciano Di Croce
09 August 2021
Marcin M. Jacak, Piotr Jóźwiak, … Janusz E. Jacak
Scientific Data volume 9, Article number: 700 (2022)
636
12
Metrics details
Research can be more transparent and collaborative by using Findable, Accessible, Interoperable, and Reusable (FAIR) principles to publish Earth and environmental science data. Reporting formats—instructions, templates, and tools for consistently formatting data within a discipline—can help make data more accessible and reusable. However, the immense diversity of data types across Earth science disciplines makes development and adoption challenging. Here, we describe 11 community reporting formats for a diverse set of Earth science (meta)data including cross-domain metadata (dataset metadata, location metadata, sample metadata), file-formatting guidelines (file-level metadata, CSV files, terrestrial model data archiving), and domain-specific reporting formats for some biological, geochemical, and hydrological data (amplicon abundance tables, leaf-level gas exchange, soil respiration, water and sediment chemistry, sensor-based hydrologic measurements). More broadly, we provide guidelines that communities can use to create new (meta)data formats that integrate with their scientific workflows. Such reporting formats have the potential to accelerate scientific discovery and predictions by making it easier for data contributors to provide (meta)data that are more interoperable and reusable.
Making Earth and environmental science data Findable, Accessible, Interoperable, and Reusable (FAIR)1,2 contributes to research that is more transparent and reproducible3. Search engines and data repositories2,4,5 have enabled advances in data preservation, findability, and accessibility. However, data interoperability and reuse remain major challenges in part due to the diversity of Earth science data, and because researchers may lack time and funding for data management or awareness of tools and resources to make data more reusable5,6. This results in barriers to scientific research and knowledge generation; for example, synthesis of data across different sources can be extremely time-consuming when data and metadata are not standardized in a common, well-defined format.
Standards for data and metadata, hereafter referred to as (meta)data standards, have been proposed as important elements to make Earth and environmental science data easier to find, understand and reuse7,8,9,10. Formal (meta)data standards are typically accredited by large governing bodies and emphasize making data broadly reusable11. For example, the International Organization for Standardization (ISO) 8601 standard provides guidelines for formatting date and timestamps and has been adopted in a wide range of research and business sectors12. The Open Geospatial Consortium’s Sensor Observation Service standard13 outlines standardized ways of pulling sensor data from web interfaces. Such accredited standards are extraordinarily useful, but are available only for a few environmental data types and can take over a decade to build governing processes and consensus.
In contrast, reporting formats are community efforts aimed at harmonizing diverse environmental data types without the oversight of the governing protocols or working groups that maintain vocabularies and extensive documentation. There are reporting formats for different research domains and data types including water quality14 and meteorological data15. Reporting formats are typically more focused within scientific domains—for example, marine observations16 or solid earth geoscience17. Reporting formats can enable efficient collection and harmonization of information needed to understand and reuse specific types of data within a research community. For example, the use of FLUXNET’s half-hourly flux and meteorological reporting format18 enables both access and reuse of consistently formatted carbon, water, and energy flux data from thousands of sampling locations across the world. However, reporting formats do not exist for most environmental data types, and even if they do, complexity and lack of resources can limit their adoption9.
There are many scientific benefits when research communities adopt reporting formats, ranging from organizing data collection in the field or lab to more efficient data reuse in synthesis and modeling efforts. Reporting formats can facilitate data sharing within a group, provide guidelines for consistent data collection, enable streamlined scientific workflows, and enable long-term preservation of knowledge that may not be typically stored or reported with the data19,20. Moreover, research disciplines are beginning to operationalize and implement practices21,22 to achieve the original FAIR guiding principles21,22. Reporting formats developed by the research communities for which they are intended are seen as a critical step toward achieving greater data interoperability and reuse22.
A variety of multidisciplinary data are generated in research sponsored by the U.S. Department of Energy (DOE) and stored in the Environmental Systems Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE) data repository4,23. Integration and analysis of diverse data types such as hydrological, geological, ecological, biological, and climatological data is an essential element of complex environmental systems science (ESS) research. However, such interdisciplinary data integration presents unique challenges, such as inconsistent use of terms, formats, and metadata across disciplines24. In this manuscript, we describe and harmonize 11 diverse and complementary (meta)data reporting formats that our interdisciplinary team developed for commonly used data types in ESS research to enable their archival following FAIR principles in general purpose repositories such as ESS-DIVE. These include guidelines to format and describe general research elements (e.g., general file metadata, tabular data, physical samples, model data), as well as guidelines developed for more specific data types relevant to interdisciplinary research (e.g., biogeochemical samples, soil respiration, leaf-level gas exchange). As part of this process, we adopted or used components of existing reporting formats or standards to the greatest extent possible, and also developed new reporting formats for some data types. These can be used individually or collectively in scientific workflows, and many of the formats are widely applicable for environmental research. Moreover, the process we used for developing the formats—including our approach to obtain community consensus, mirror documentation across several web platforms, and track community feedback—can be used by other research communities to develop reporting formats for their own purposes.
Our community-centric approach to developing reporting formats had four key outcomes that are broadly important to making scientific data more reusable. First, the teams reviewed a total of 112 pre-existing data standards and other data resources (data repositories, data systems, datasets, projects) to create (meta)data crosswalks (Supplementary Files 1–20). Such crosswalks provide a tabular map of existing resources related to each data type, allowing the teams to identify gaps in existing standards, and determine which variables, terms, and metadata were essential to harmonize and incorporate into their reporting formats. At the onset of the review process, ESS-DIVE recommended adopting existing standards to the extent possible. However, we found that for all 11 data types, none entirely met ESS research community needs, and this necessitated development of all 11 reporting formats.
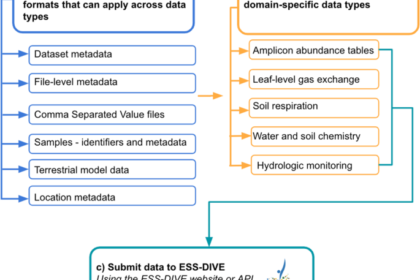
Second, we created 11 reporting formats (Supplementary Table 1) that encompass a range of complex and diverse ESS (meta)data fields that can be used when researchers upload data to ESS-DIVE. Six of the reporting formats created by our community of scientists are cross-domain reporting formats (Fig. 1a), which apply broadly to data across different scientific disciplines. These reporting formats were developed to help researchers more consistently format their (meta)data for interdisciplinary science applications and include basic dataset metadata for citation and findability25, file-level metadata26, guidelines for formatting comma separated value (CSV) files27, sample metadata28, terrestrial model data archiving guidelines29, and research locations metadata30. The remaining five reporting formats apply to different domain data types (Fig. 1b) and include microbial amplicon abundance tables31, leaf-level gas exchange32, soil respiration33, sample-based water and soil chemistry measurements34, and water level and sonde-based hydrologic measurements35. All reporting formats have a minimal set of required metadata fields necessary for programmatic data parsing and optional fields that provide detailed spatial/temporal context about the sample useful to downstream scientific analyses. Throughout development, we aimed to strike a balance between pragmatism for the scientists reporting data and machine-actionability that is emblematic of FAIR data. A comparison between FAIR guiding principles and our reporting formats (Supplementary Table 2) highlights how a community-centric effort like ours can move data archiving towards achieving many FAIR data principles (though see discussion for limitations).
Workflow to help determine which (meta)data reporting formats apply to datasets. The set of 11 ESS-DIVE (meta)data formats are either (a) cross-domain guidelines that can be applied to many data types or (b) are data type-specific. For those archiving data with ESS-DIVE, researchers can upload data through the ESS-DIVE web user interface155 or programmatically through an API.
Together, these 11 reporting formats are part of a flexible, modular, and integrated framework (Fig. 1) that can accommodate new reporting formats in the future, and enable their findability and accessibility individually or collectively. As part of the framework development, all teams created templates with harmonized terms and formats to be internally consistent as much as possible. For example, dates are always reported in YYYY-MM-DD format. Whenever reporting formats include spatial data, the variables are harmonized as “latitude” and “longitude” and reported in decimal degrees with common bounds (−90 to 90 and −180 to 180, respectively). All formats that require CSV files adopted as many recommendations from the CSV reporting format as possible. Data collected using the water and soil chemistry, and amplicon reporting formats have an option to report a persistent identifier for associated samples [International Generic Sample Number (IGSN)], to enable effective tracking across online data systems, as outlined in the Sample ID reporting format.
The third outcome is related to how we shared and archived all reporting formats in three ways, each with a distinct use. First, all reporting formats are published as datasets in the ESS-DIVE repository, which enables direct, public download and citation upon use. Second, each reporting format is hosted on the version control platform GitHub, which enables ongoing edits and versioning while also allowing users to provide feedback36. Third, the most up-to-date reporting format content from GitHub is rendered as a project website through the service GitBook37. We mirrored the reporting format instructions and templates across several web platforms to ensure the documentation is available in a variety of digital formats to serve the needs of various user groups and stakeholders. GitHub is likely a more familiar platform and user interface for software engineers and informatics specialists, for example, while GitBook websites may be preferred by Earth science researchers.
Lastly, we formulated guidelines (Box 1) for research communities that want to replicate our model of community-centric (meta)data reporting format development. We encourage (1) reviewing existing standards, (2) developing a crosswalk of terms across relevant standards or ontologies of interest, (3) iteratively developing templates and documentation with feedback from prospective users, (4) assembling a minimum set of (meta)data required for reuse, and (5) hosting finalized documentation on platforms that can be publicly accessed and updated easily.
1. Research existing (meta)data standards and other data resources across agencies and organizations both within the US and internationally.
2. Create a (meta)data crosswalk (Supplementary Files 1–10) to define how other standards and data resources translate to the proposed reporting format.
3. Work with the scientific community to iteratively develop and obtain feedback (see Fig. 2) on (meta)data reporting format.
4. Develop documentation (instructions, templates, variables, descriptions, units, metadata) to support the format. Consider appropriate file formats for any templates.
5. Archive finalized version of the reporting format in a long-term data repository as well as a version control platform (e.g., GitHub37).
Many scientific journals and funders require data deposition in long-term repositories. However, in many cases, data are submitted to repositories in bespoke formats with little (meta)data standardization5. Community-led (meta)data reporting formats like the set described in this paper can enable archived data to be more reusable and interoperable21,22. Our scientist-centric approach to creating the formats helped to determine workflows that are most useful and practical for researchers to adopt. Here we discuss important aspects that need to be considered in development and use of such reporting formats.
Reporting formats can help researchers organize and synthesize their own (meta)data for their research purposes. It can be challenging for small teams, or even individuals to keep track of data collected over multi-year field campaigns or laboratory experiments19,20. Early adoption of a consistent way of compiling data can help individuals or research teams avoid ad hoc data collection practices and also help researchers efficiently integrate their data, particularly when multiple analyses or teams are involved.
Moreover, community reporting formats can lead to greater data accessibility and reuse. For example, researchers in the Ameriflux network38 organize flux data in the Flux Processing (FP-in) reporting format18. When participants in the network agree to provide their flux data in this format39, benefits include: 1) access to data services such as automated QA/QC of datasets and value-added ONEFlux data processing40, 2) Digital Object Identifier assignment which helps to track dataset citation and reuse, and 3) potential to increase findability of their data. Similarly, when contributors upload datasets on ESS-DIVE, they are offered automated metadata quality assessments, and published data are assigned DOIs and made searchable across the DataONE network. In another example, the Watershed Function Scientific Focus Area project41 adopted ESS-DIVE’s water and soil chemistry reporting format as an initial step towards establishing a field data workflow in a community observatory where diverse hydrological, geochemical, geophysical, ecological, and remote sensing datasets are collected42. The use of the reporting format will make it possible for researchers to synthesize data on chemical concentrations both within and across field locations.
Application of the reporting formats also allows for the use of tools and services that enhance data curation, findability and reuse. As an example, some of the fields in ESS-DIVE’s dataset metadata reporting format25 allow programmatic metadata quality validation, which checks for field presence, format, and length. Because these metadata can be mapped to a variety of machine-readable metadata formats including JSON-LD and the U.S. Department of Energy’s Office of Scientific and Technical Information (OSTI) reporting formats43. This further enabled transforming and disseminating ESS-DIVE datasets across other platforms such as Google Dataset Search, DataONE, OSTI and DataCite.
The development of these reporting formats was driven by the scientific need for practical tools for data management, while improving the potential for data reuse achieving many of the FAIR guiding principles (Supplementary Table 2). We made several pragmatic choices to ensure that the reporting formats would have a low barrier to adoption by time-limited researchers. This included investigating whether using pre-existing reporting formats “off the shelf” would meet project and researcher’s scientific needs and workflows. Although it is desirable to use existing formats whenever possible, we found that there were many circumstances when they do not directly apply to a scientific community’s research (meta)data needs. For example, although the Water Quality Exchange format14 is used within the United States to report water quality monitoring data by local, state, and federal agencies, the format was not entirely suitable for ESS-DIVE’s purposes. Some of the concerns raised by the community included: 1) the structure of the data and metadata templates that are used for regulatory reporting were considered to be cumbersome and inefficient for scientific use (e.g., containing redundant elements of sampling and analytical methodology along with the data) and 2) the required vocabularies (as specified in the template dictionary) were found to be difficult to use because they included several terms that were unnecessary, while missing terms for specific analytes of interest to the community.
To address these concerns, we developed the ESS-DIVE reporting format for sample-based water and soil chemistry34 that is more suitable for files typically generated in scientific laboratories. It borrows elements from the WQX standard, but provides flexibility in format and terminology, while capturing sufficient metadata and vocabularies to enable data exploration and reuse including the ability to use scripts to compare and combine different datasets34. In this way, the water and soil chemistry reporting format achieves some component of FAIR guiding principle “I2” that suggests using ontologies, while still being responsive to a research community that desired flexibility in research terminology (Supplementary Table 2).
Similarly, when creating the sample ID metadata reporting format, we decided to extend the existing IGSN sample identifier template and guidelines in ESS-DIVE’s Sample ID reporting format to meet researchers’ need to link interdisciplinary environmental and biological samples, and to minimize effort in providing information for sample collections44. In this case, incorporating IGSNs ensures that researchers using this format achieve FAIR principle “F3” and have globally unique identifiers for their data products, which facilitates tracking associated sample data across multiple online data systems. In an effort to be pragmatic, we decided to lower the threshold for adoption of the sample ID reporting format (and nearly all others; Supplementary Table 2) by compromising on elements that would achieve FAIR principle “I3” related to machine readable knowledge representation. All reporting formats encourage users to define variables in a data dictionary. Though this may not be fully machine readable according to the FAIR principles21, this method of defining variables is a key step toward reusable and machine actionable data. The feedback gathered when creating our Sample ID reporting format was then provided to the broader IGSN community to help improve the IGSN metadata template for interdisciplinary science45,46.
Through the process, we learned that many (meta)data standards are not accessible to a typical researcher and require a significant learning curve to become fluent in the informatics terminology used by established data standards. For example, the Open Geospatial Consortium’s data standard for environmental sensors13 is a detailed schema described over 100 pages, which is challenging for a typical scientific researcher to understand and implement. Hence, we had to make several pragmatic choices to ensure that the reporting formats would be amenable to adoption by time-limited researchers. Once choice involved replacing terms in existing standards with words that were more intuitive to scientists. For example, whilst there was no reporting format for leaf-level gas exchange data, a crosswalk of the instrument output from a relatively small number of instrument manufacturers quickly identified a common terminology that already had broad acceptance and use by the scientific community (Supplementary File 7). By using crosswalks (Supplementary Files 1–10) our teams were able to map ESS-DIVE’s reporting formats to many existing (meta)data standards and other data resources, and, in the future, will allow building tools that enable interoperability with different systems. We also simplified the reporting format templates and instructions to the greatest extent possible by specifying a few required fields and several more optional fields to provide additional details.
Our model and guidelines of supporting and empowering the scientific community to develop (meta)data reporting formats that meet their needs can enable other communities to undertake these internal data standardization efforts that make their data even more useful beyond the purpose for which they were collected (Box 1). We acknowledge that other research infrastructures have made important strides toward data standardization within research communities though they can still take dozens of years to manifest17. We found value in including a broad range of stakeholders in the process, and included field personnel who make the measurements, instrument manufacturers, and scientists who use the data in models or synthesis activities47.
There are incentives that can help promote widespread adoption of these or other formats to justify the time investment required for individual researchers or teams into scientific workflows. First, involving data collectors and reusers at the core of the development process makes the resulting formats more pragmatic and scientifically useful. Importantly, the domain scientists involved in the reporting format development became community ambassadors and helped engage their use by fellow researchers through conference presentations and peer-reviewed papers44,47,48,49. Second, we expanded our user community by sharing information about the reporting formats through a series of webinars, documentation, tutorials, and personalized community outreach. These incentives have had some success, as evidenced by the datasets submitted to ESS-DIVE using one or more of the reporting formats within a few months after they were finalized (Table 1).
We identify some future work that can potentially lower the barrier to adopting reporting formats, provide added benefits to those who use the formats, and make (meta)data FAIRer10. Currently, ESS-DIVE applies a set of manual checks to datasets uploaded to ESS-DIVE that follow the reporting format. However, development of automated formatting checkers50 would help users instantly validate their datasets against reporting format guidelines. Other types of software can also be built around the reporting formats. For example, software could be developed to automatically convert sensor or instrument-derived data into the units requested by a reporting format. As a starting point for this work, the file-level metadata reporting format already includes an open-source script51 that enables reading and parsing data files submitted in that format. The leaf-level gas exchange reporting format includes a detailed translation table matching the reporting format data variables with standard outputs from 10 commonly used, commercially produced instruments. This could provide the foundation for development of conversion software to automatically format data with the recommended variable names and units. ESS-DIVE is also planning a data integration and fusion component of the repository that will facilitate synthesizing and analyzing datasets that adhere to any of the 11 ESS-DIVE reporting formats. Enabling advanced queries within the files will require development of software and data parsers so that a great number of reporting formats achieve FAIR principle “F4” which calls for data to be fully searchable.
With more data being generated than ever, reusable data can have substantial societal, economic, and scientific impacts. But for Earth and environmental science data, which are complex and heterogeneous, achieving reusability will require concentrated effort at (meta)data standardization within research communities. Our work to develop 11 community (meta)data reporting formats is a critical step to making Earth and environmental science data more reusable because we emphasize human readability that is compatible with machine readability. We hope that our model of empowering research communities to self-organize and create their own (meta)data reporting formats will enable other communities to undertake these internal data standardization efforts that make their data even more useful beyond the purpose for which they were collected.
Earth and environmental science data are complex, multi-scale, and span diverse research domains such as geology, hydrology, climate, ecology, and biology. At ESS-DIVE, we initiated a community-centric model that engaged domain scientists to develop formats for common Earth science data types. The objective was to create formatting guidelines and templates that would gather the minimum but sufficient metadata or data necessary for data interpretation and reuse.
Each team conducted a review of existing standards (Supplementary Table 1), involving both literature searches and exploring resources from informatics groups (e.g., Research Data Alliance and Earth Science Information Partners) or agencies working with similar data, to identify whether any existing data standards or conventions could be used directly or to inform their reporting format. Based on this review, each team created tabular ‘crosswalks’ (Supplementary Files 1–10) to map related terminology from relevant standards. This process helped identify gaps in existing standards, and determine important elements that had to be present, and variations in terminology used across different standards that required harmonization. For example, some existing standards report date and time under the column name ‘datetime’ while another reports the same information, as ‘ValueDateTime’ (see example of a terminology crosswalk35). Here, we provide a brief narrative of methods for each reporting format with details on existing data standards and other data resources reviewed during reporting format development. For further details on the technical aspects of each reporting format, please refer to ESS-DIVE’s community space on GitHub36 or view the datasets for each reporting format submitted to ESS-DIVE (Supplementary Table 1).
Each team created instructions and (meta)data templates for their reporting formats. The teams piloted the formats within their research groups and communities to ensure the templates were practical and useful for scientists who collect and reuse data (Fig. 2). In total, 247 individuals representing 128 institutions provided input at various stages of the reporting format development process. As the reporting format instructions and templates reached a final stage, they published the “ready-to-use” reporting formats in three locations each with distinct benefits for the end-users: GitHub37, GitBook, and the ESS-DIVE data repository to enable findability and long-term preservation.
Each of the 11 ESS-DIVE (meta)data reporting formats were developed in cross-functional teams that often involved domain scientists, software engineers, and informatics specialists.
The goal for creating the dataset metadata reporting format was to ensure that any dataset submitted to ESS-DIVE would have complete and descriptive metadata to enable its findability and citation upon use. The ESS-DIVE team reviewed machine and human-readable metadata standards including the Ecological Metadata Language52 as well as JSON for Linking Data53. The ESS-DIVE metadata reporting format follows existing metadata standards as much as possible (e.g., ‘title’ in Ecological Metadata Language is also ‘title’ for ESS-DIVE’s metadata).
The file-level metadata reporting format was developed for users to provide details about the individual files contained within a dataset. The review of existing standards26 included file-level metadata used across 6 organizations (e.g., USGS, NEON).
The CSV reporting format was developed to provide guidelines for more consistently formatting tabular data27. The intention was to make this a domain agnostic set of guidelines so that anyone who works with tabular data can use the format in their research to make tabular data more interoperable and machine-readable. The team reviewed existing standards and guidelines (Supplementary Table 1) including recommendations from the Environmental Data Initiative (e.g., do not mix data types in a column) and the ORNL DAAC (e.g., indicating missing numeric values with −9999).
The ESS-DIVE Sample ID reporting format28 aligns as much as possible with extensive work on IGSN54 with the goal of standardizing sample collection metadata and more efficiently tracking physical samples sent to different collaborators, labs, data systems, etc. This work also reviewed 12 different standards and data resources to provide recommendations for improving interoperability of biological8,55 and environmental samples14.
The model data archiving reporting format29 was informed by input from the DOE’s land modeling community and other guidelines from the American Geophysical Union and National Science Foundation Earthcube communities. In developing the guidelines49, the goal was to help modelers make decisions about which components of their terrestrial models should be archived in a long-term data repository. The guidelines were developed with input on which model data were most useful to archive, how long they remained useful, and what scientific purpose they would serve.
The goal of developing the location metadata reporting format was to provide generalized guidelines for describing locations used in research. The review of existing standards included metadata templates from specific projects at some of the DOE’s National Labs to understand the different field sampling strategies of large interdisciplinary projects. The review also included known standards and guidelines for recording locations such as Climate and Forecast Conventions56, the Federal Geographic Data Committee’s Content Standard for Digital Geospatial Metadata57 and the Open Geospatial Consortium58.
In addition to the set of 6 cross-domain reporting formats described above, we also developed 5 formats that are tailored to specific data types commonly used in the terrestrial and subsurface ecosystem research community. ESS-DIVE’s goal was to engage Earth and environmental scientists to determine practical reporting formats that data contributors are willing to use while at the same time ensuring a high potential for data reuse.
The reporting format for amplicon abundance table metadata was developed to facilitate consistent reporting of microbiome sample data with the format of these tables following ESS-DIVE’s CSV file guidelines. Required data (e.g., representative sequences) were chosen to support comparisons of abundance tables across studies. The reporting format distinguishes between sequencing metadata and bioinformatic processing metadata for amplicon abundance tables. As much as possible, the team aligned recommendations for sequencing metadata with the existing standards developed by the Genomic Standards Consortium for minimum information about a marker gene sequence and minimum information about any (x) sequence55 (Supplementary File 6). In the absence of an existing standard for bioinformatic processing metadata, the reporting format contains a minimal set of fields to capture the data processing steps most relevant for comparing and combining amplicon counts across studies (Supplementary Table 1). The final set of sequencing and bioinformatic metadata fields selected were informed by a community of scientists involved with either the development of microbiome data pipelines or conducting microbiome studies in both field and lab settings.
The team working on this reporting format32 reviewed existing conventions used in plant trait databases, large data collections developed for synthesis papers, and the variable descriptions that are part of standard instrument outputs in order to determine the most suitable variable names to use to report leaf-level gas exchange data. Templates for formatting metadata about the methods and sample materials used in an experiment, as well as details on the instrumentation involved in collecting data were developed through an iterative process of input and review open to all interested stakeholders. The reporting format is designed to be flexible and modular, provides guidelines on the archive of raw and processed data, and seeks to capture experimental metadata needed to interpret and reuse these data types47.
To create the soil respiration reporting format, this team reviewed and integrated recommendations from 9 existing guidelines and standards (Supplementary Table 1)33. The review captured an array of how different standards format their general metadata and data (e.g., formatting date and timestamps) and also accounted for a range of soil-atmosphere gas exchange data types (e.g., GHGs or radiocarbon)48.
The goal in creating a reporting format for water-soil-sediment data was to harmonize chemical concentration data that span several measurement types. The review included 15 standards (Supplementary Table 1) for related environmental chemistry measurements including metadata elements from the EPA’s WQX14 as well as EarthChem59. Based on input from the potential ESS user community that included both data collectors, managers, and modelers, we developed a reporting format based on community input34.
This reporting format harmonizes variables common to sonde-based hydrologic monitoring research including water level, temperature, and pH data. The existing standards and/or data sources included in the crosswalk for the hydrologic monitoring reporting format (Supplementary Table 1) were chosen for inclusion given their common use in the scientific community. They aligned generally on the types of hydrologic metadata to record (e.g., information about dates and times as well as information about data collection sites) but had different terminology across each of the resources35. The development of the reporting format included a review of additional data sources and standards beyond those listed in the crosswalk (Supplementary Table 1).
Each data reporting format and all supporting documentation are hosted on our GitHub Community Space36 and archived in the ESS-DIVE data repository25,26,27,28,29,30,31,32,33,34,35. The supplementary information for this manuscript is also archived in ESS-DIVE60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147.
We have made code available which enables file-level metadata extraction51 for files that adhere to the reporting format.
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Article PubMed PubMed Central Google Scholar
Stall, S. et al. Advancing FAIR data in Earth, space, and environmental science. Eos 99 (2018).
Toelch, U. & Ostwald, D. Digital open science-Teaching digital tools for reproducible and transparent research. PLoS Biol. 16, e2006022 (2018).
Article PubMed PubMed Central Google Scholar
Varadharajan, C. et al. Launching an Accessible Archive of Environmental Data. Eos 100 (2019).
Tenopir, C. et al. Data sharing, management, use, and reuse: Practices and perceptions of scientists worldwide. PLoS One 15, e0229003 (2020).
Article CAS PubMed PubMed Central Google Scholar
Perrier, L., Blondal, E. & MacDonald, H. The views, perspectives, and experiences of academic researchers with data sharing and reuse: A meta-synthesis. PLoS One 15, e0229182 (2020).
Article CAS PubMed PubMed Central Google Scholar
Sansone, S. A. et al. FAIRsharing as a community approach to standards, repositories and policies. Nat. Biotechnol. 37, 358–367 (2019).
Article CAS PubMed PubMed Central Google Scholar
Wieczorek, J. et al. Darwin Core: an evolving community-developed biodiversity data standard. PLoS One 7, e29715 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Poisot, T., Bruneau, A., Gonzalez, A., Gravel, D. & Peres-Neto, P. Ecological Data Should Not Be So Hard to Find and Reuse. Trends Ecol. Evol. 34 (2019).
Michener, W. K. Ecological data sharing. Ecol. Inform. 29, 33–44 (2015).
Article Google Scholar
Lin, D. et al. The TRUST Principles for digital repositories. Sci. Data 7, 144 (2020).
Article PubMed PubMed Central Google Scholar
ISO. Date and Time Format (ISO Standard Number 8601-1:2019) (2019).
Bröring, A., Stasch, C. & Echterhoff, J. OGC Sensor Observation Service Interface Standard, Version 2.0. (2012).
Read, E. K. et al. Water quality data for national‐scale aquatic research: The Water Quality Portal. Water Resour. Res. 53, 1735–1745 (2017).
Article ADS Google Scholar
AmeriFlux. BADM: Biological, Ancillary, Disturbance, and Metadata https://ameriflux.lbl.gov/data/badm/ (2020).
Dañobeitia, J. J. et al. Toward a Comprehensive and Integrated Strategy of the European Marine Research Infrastructures for Ocean Observations. Front. in Mar. Sci. 7 (2020).
Cocco, M. et al. The EPOS Research Infrastructure: a federated approach to integrate solid Earth science data and services. Ann. Geophys. 65, DM208–DM208 (2022).
Article Google Scholar
Flux Processing (FP-In). Half-Hourly/Hourly Data Upload Format https://ameriflux.lbl.gov/half-hourly-hourly-data-upload-format/ (2017).
Goodman, A. et al. Ten Simple Rules for the Care and Feeding of Scientific Data. PLoS Comput. Biol. 10, e1003542 (2014).
Article PubMed PubMed Central Google Scholar
Lowndes, J. S. S. et al. Our path to better science in less time using open data science tools. Nat. Ecol. Evol. 1, 0160 (2017).
Article Google Scholar
Jacobsen, A. et al. FAIR principles: Interpretations and implementation considerations. Data Intelligence 2, 10–29 (2020).
Article Google Scholar
Bailo, D. et al. Perspectives on the Implementation of FAIR Principles in Solid Earth Research Infrastructures. Front. Earth Sci. Chin. 8 (2020).
Environmental System Science Data Infrastructure for a Virtual Ecosystem. ESS-DIVE https://data.ess-dive.lbl.gov/data.
Hills, D. J. et al. Earth and space science informatics perspectives on integrated, coordinated, open, networked (ICON) science. Earth Space Sci. 9 (2022).
Agarwal, D. et al. ESS-DIVE Reporting Format for Dataset Package Metadata. ESS-DIVE https://doi.org/10.15485/1866026 (2022).
Velliquette, T. et al. ESS-DIVE Reporting Format for File-level Metadata. ESS-DIVE https://doi.org/10.15485/1734840 (2021).
Velliquette, T. et al. ESS-DIVE Reporting Format for Comma-separated Values (CSV) File Structure. ESS-DIVE https://doi.org/10.15485/1734841 (2021).
Damerow, J. et al. ESS-DIVE global sample numbers and metadata reporting format for Environmental Systems Science (IGSN-ESS). ESS-DIVE https://doi.org/10.15485/1660470 (2020).
Simmonds, M. B. et al. ESS-DIVE guidelines for archiving terrestrial model data. ESS-DIVE https://doi.org/10.15485/1813868 (2021).
Crystal-Ornelas, R. et al. ESS-DIVE Reporting Format for Location Metadata. ESS-DIVE https://doi.org/10.15485/1865730 (2022).
Weisenhorn, P. & Beilsmith, K. ESS-DIVE Reporting Format for Amplicon Abundance Tables. ESS-DIVE https://doi.org/10.15485/1865729 (2022).
Ely, K. S., Rogers, A. & Crystal-Ornelas, R. ESS-DIVE reporting format for leaf-level gas exchange data and metadata. ESS-DIVE https://doi.org/10.15485/1659484 (2020).
Bond-Lamberty, B., Christianson, D. S., Crystal-Ornelas, R., Mathes, K. & Pennington, S. C. ESS-DIVE reporting format for field measurements of soil respiration. ESS-DIVE https://doi.org/10.15485/1798520 (2021).
Boye, K. et al. ESS-DIVE Reporting Format for Sample-based Water and Soil Chemistry Measurements. ESS-DIVE https://doi.org/10.15485/1865731 (2022).
Goldman, A. E., Ren, H., Torgeson, J. & Zhou, H. ESS-DIVE Reporting Format for Hydrologic Monitoring Data and Metadata. ESS-DIVE https://doi.org/10.15485/1822940 (2021).
ESS-DIVE Community Space. ESS-DIVE Community Space https://github.com/ess-dive-community (2021).
Crystal-Ornelas, R. et al. A guide to using GitHub for developing and versioning data standards and reporting formats. Earth Space Sci. 8 (2021).
Novick, K. A. et al. The AmeriFlux network: A coalition of the willing. Agric. For. Meteorol. 249, 444–456 (2018).
Article ADS Google Scholar
AmeriFlux Data Policy. AmeriFlux Data Policy https://ameriflux.lbl.gov/data/data-policy/ (2021).
Onboarding and Orientation for new site teams. https://ameriflux.lbl.gov/sites/onboarding-and-orientation-for-new-site-teams/ (2017).
Hubbard, S. S. et al. The East River, Colorado, watershed: A mountainous community testbed for improving predictive understanding of multiscale hydrological–biogeochemical dynamics. Vadose Zone J. 17, 1–25 (2018).
Article Google Scholar
Kakalia, Z. et al. The Colorado East River community observatory data collection. Hydrol. Process. 35 (2021).
OSTI. Instructions for announcement of U.S. Department of Energy (DOE) publicly available scientific research datasets https://www.osti.gov/elink/F2416instruct.jsp (2017).
Damerow, J. et al. Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences. Data Sci. J. 20, 1–19 (2021).
Article Google Scholar
Klump, J. et al. Towards globally unique identification of physical samples: Governance and technical implementation of the IGSN global sample number. Data Sci. J. 20 (2021).
Richard, S. M. et al. Internet of samples. Proc. Assoc. Inf. Sci. Technol. 58, 813–815 (2021).
Article Google Scholar
Ely, K. S. et al. A reporting format for leaf-level gas exchange data and metadata. Ecol. Inform. (2021).
Bond-Lamberty, B., Christianson, D. S., Crystal-Ornelas, R., Mathes, K. & Pennington, S. C. A reporting format for field measurements of soil respiration. Ecol. Inform. (2021).
Simmonds, M. B. et al. Guidelines for Publicly Archiving Terrestrial Model Data to Enhance Usability, Intercomparison, and Synthesis. Data Sci. J. 21 (2022).
Fowler, D., Barratt, J. & Walsh, P. Frictionless data: Making research data quality visible. Int. J. Digit. Curation 12, 274–285 (2018).
Article Google Scholar
McNelis, J., Crow, M. & Devarakonda, R. ESS-DIVE File Level Metadata Extractor. DOE Code https://doi.org/10.11578/DC.20201103.5 (2020).
Jones, M. et al. Ecological Metadata Language version 2.2.0. KNB Data Repository https://doi.org/10.5063/F11834T2 (2019).
Sporny, M., Longley, D., Kellogg, G., Lanthaler, M. & Lindström, N. JSON-LD 1.0. W3C recommendation 16, 41 (2014).
Google Scholar
Lehnert, K. A., Klump, J., Wyborn, L. & Ramdeen, S. IGSN: Trustworthy and Sustainable Services for FAIR Samples. In Geophysical Research Abstracts 21 (2019).
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
Article CAS PubMed PubMed Central Google Scholar
CF Conventions and Metadata. https://cfconventions.org/index.html (2020).
Federal Geographic Data Committee. Content Standard for Digital Geospatial Metadata. https://ci.nii.ac.jp/naid/10016800076/ (1998).
Observations and Measurements. Observations and Measurements https://www.ogc.org/standards/om (2011).
Walker, J. D., Lehnert, K. A., Hofmann, A. W., Sarbas, B. & Carlson, R. W. EarthChem: International Collaboration for Solid Earth Geochemistry in Geoinformatics. in vol. 2005 IN44A-03 (2005).
Crystal-Ornelas, R. et al. Data from: “Enabling FAIR data in Earth and environmental science with community-centric (meta)data reporting formats”. ESS-DIVE https://doi.org/10.15485/1866606 (2022).
JSON for Linking Data. JSON for Linking Data https://json-ld.org/ (2016).
Brase, J. DataCite – A Global Registration Agency for Research Data. In 2009 Fourth International Conference on Cooperation and Promotion of Information Resources in Science and Technology 257–261 (2009).
Evans, K. et al. ASCII File Format Guidelines for Earth Science Data. (2016).
Federal Geographic Data Committee. National Geospatial Data Assets (NGDA) Metadata Guidelines. (2016).
Shafranovich, Y. Common Format and MIME Type for Comma-Separated Values (CSV) Files. (2005).
USGS Data Dictionaries. https://www.usgs.gov/data-management/data-dictionaries (2020).
EDI. Five phases of data publishing – Phase 2: Format and QC data tables https://environmentaldatainitiative.org/five-phases-of-data-publishing/phase-2/ (2019).
Pepler, S. & Parton, G. BADC-CSV Format for Data Exchange. https://help.ceda.ac.uk/article/105-badc-csv (2009).
Hsu, L. How “clean” should an Excel file be to be considered machine readable. https://my.usgs.gov/confluence/pages/viewpage.action?pageId=559852026 (2016).
NEON. NEON file naming conventions. https://data.neonscience.org/file-naming-conventions (2022).
Tarboton, D. G., Horsburgh, J. S. & Maidment, D. R. CUAHSI community Observations Data Model (ODM) version 1.1 design specifications. Des Doc (2008).
StreamPulse uploading data. http://pulseofstreams.weebly.com/uploading_data.html (2022).
Cerf, V. ASCII format for Network Interchange. (1969).
Newell, D. B. & Tiesinga, E. The International System of Units (SI) (2019 Edition). (National Institute of Standards and Technology, 2019).
EPSG. WGS 84. (1984).
ORNL DAAC CSV Standards. https://daac.ornl.gov/submit/csvstandards/ (2018).
Data Quality Review Checklist. https://daac.ornl.gov/submit/qachecklist/https://daac.ornl.gov/submit/qachecklist/ (2022).
USGS Data Templates. https://www.usgs.gov/products/data-and-tools/data-management/data-templates (2022).
National Archives. https://www.archives.gov/records-mgmt/policy/transfer-guidance-tables.html#structuredata (2022).
Klyne, G. & Newman, C. Date and Time on the Internet: Timestamps. (2002).
Loescher, H. AmeriFlux BASE CR-Lse La Selva. AmeriFlux https://doi.org/10.17190/AMF/1246013 (2016).
Torn, M. & Dengel, S. AmeriFlux US-NGB NGEE Barrow. AmeriFlux https://doi.org/10.17190/AMF/1436326 (2018).
Brokaw, N. Luquillo Forest Dynamics Plot (LFDP) Liana Data. Environmental Data Initiative https://doi.org/10.6073/PASTA/EABE6E15723324EA3938B456D5BB02C2 (2017).
Bueno de Mesquita, C. P. Plant colonization of moss-dominated soils in the alpine: Microbial and biogeochemical implications. Environmental Data Initiative https://doi.org/10.6073/PASTA/C0CACD100CD89DA258B40A77FBB2FD4C (2019).
Brooks, S. East Fork Poplar Creek sonde data at kilometer 16.2 water year 2019. Oak Ridge National Laboratory (ORNL) https://doi.org/10.12769/1569821 (2019).
Riscassi, A. & Brooks, S. East Fork Poplar Creek discharge at kilometer 5.4 water year 2012. Oak Ridge National Laboratory (ORNL) https://doi.org/10.12769/1489524 (2019).
National Ecological Observatory Network (NEON). Coarse downed wood bulk density sampling (DP1.10014.001). National Ecological Observatory Network (NEON) https://doi.org/10.48443/Z0TG-QS14 (2020).
Salmon, V., Iversen, C., Childs, J. & VanderStel, H. NGEE arctic plant traits: Soil cores, Kougarok road mile marker 64, Seward peninsula, Alaska, 2016. NGEE Arctic, Oak Ridge National Laboratory (ORNL) https://doi.org/10.5440/1346200 (2019).
Philben, M. et al. Results of experimental additions of organic nitrogen on soil organic matter decomposition, teller road site, Seward peninsula, 2017 and 2018. NGEE Arctic, Oak Ridge National Laboratory (ORNL) https://doi.org/10.5440/1454263 (2019).
Yaffar, D., Lugo, A. E., Silver, W., Cuevas, E. & Molina Colon, S. Plant root trait measurements raw data, 1962–2018, Island of Puerto Rico. Next-Generation Ecosystem Experiments Tropics; Oak Ridge National Laboratory https://doi.org/10.15486/NGT/1558773 (2019).
Norby, R. et al. Root-soil depth profile in Luquillo Experimental Forest, Puerto Rico, February, 2019. Next-Generation Ecosystem Experiments Tropics; Oak Ridge National Laboratory https://doi.org/10.15486/NGT/1574087 (2019).
Griffiths, N. & Sebestyen, S. SPRUCE porewater chemistry data for experimental plots beginning in 2013. Oak Ridge National Lab’s Terrestrial Ecosystem Science Scientific Focus Area (ORNL TES SFA) https://doi.org/10.3334/CDIAC/SPRUCE.028 (2016).
McPartland, M. Y. et al. SPRUCE: NDVI data from selected SPRUCE experimental plots, 2016–2018. Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States) https://doi.org/10.25581/SPRUCE.057/1490190 (2019).
Croteau, M. N., Sikder, M., Poulin, B. A. & Baalousha, M. Laboratory data to assess the effect of nanoparticle size and natural organic matter composition on the bioavailability of platinum nanoparticles to a model freshwater invertebrate species. U.S. Geological Survey https://doi.org/10.5066/P9G18URX (2020).
Danczak, R. E. et al. WHONDRS 48 Hour Diel Cycling Study at the Altamaha River in Georgia, USA. Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS); ESS-DIVE https://doi.org/10.15485/1577263 (2019).
Stegen, J. C. et al. WHONDRS 48 hour Diel cycling study at HJ Andrews Experimental Forest Watershed 1 (WS1). Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS); ESS-DIVE https://doi.org/10.15485/1509695 (2019).
Article Google Scholar
SESAR. SESAR Batch Registration Quick Guide. Zenodo https://doi.org/10.5281/zenodo.3874923 (2020).
IGSN Descriptive Metadata Schema. IGSN metadata https://github.com/IGSN/metadata (2017).
Weibel, S., Kunze, J., Lagoze, C. & Wolf, M. Dublin core metadata for resource discovery. Internet Engineering Task Force RFC 2413, 132 (1998).
Google Scholar
ISO 19156:2011. Geographic information — Observations and measurements https://www.iso.org/standard/32574.html (2011).
Joint Genome Institute Genome Online Database. https://genome.jgi.doe.gov/portal/ (2022).
USGS National Digital Catalog. https://www.usgs.gov/programs/national-geological-and-geophysical-data-preservation-program/national-digital-catalog (2022).
Geologic Materials Repository Working Group. The U.S. Geological Survey Geologic Collections Management System (GCMS)—A master catalog and collections management plan for U.S. Geological Survey geologic samples and sample collections. (2015).
NEON Biorepository Data Portal. https://biorepo.neonscience.org/portal/ (2022).
Hanson, B. Data policies and practices for AGU publications for models and model output. (2020).
Williams, D. N., Lawrence, B. N., Lautenschlager, M., Middleton, D. & Balaji, V. The Earth System Grid Federation: Delivering globally accessible petascale data for CMIP5. Proceedings of the Asia-Pacific Advanced Network 32, 121 (2011).
Article Google Scholar
Dryad. https://datadryad.org/ (2022).
Zenodo. https://zenodo.org/ (2022).
DAAC. https://earthdata.nasa.gov/eosdis/daacs (2021).
NCAR. https://rda.ucar.edu/ (2022).
EOL. https://www.eol.ucar.edu/about-eol (2022).
Arctic Data Center. https://arcticdata.io/ (2016).
Federal Geographic Data Committee. Content Standard for Digital Geospatial Metadata Part 1: Biological Data Profile. (1999).
Christianson, D. S. et al. A metadata reporting framework (FRAMES) for synthesis of ecohydrological observations. Ecol. Inform. 42, 148–158 (2017).
Article Google Scholar
USGS. https://dashboard.waterdata.usgs.gov/ (2022).
SNOTEL. https://wcc.sc.egov.usda.gov/reportGenerator/ (2022).
Earth Microbiome Project. https://earthmicrobiome.org/protocols-and-standards/metadata-guide/ (2022).
National Center for Biotechnology Information. SRA Metadata and Submission Overview. https://www.ncbi.nlm.nih.gov/sra/docs/submitmeta/ (2019).
Kattge, J. et al. TRY plant trait database – enhanced coverage and open access. Glob. Chang. Biol. 26, 119–188 (2020).
Article ADS PubMed Google Scholar
LeBauer, D. et al. BETYdb: a yield, trait, and ecosystem service database applied to second generation bioenergy feedstock production. Glob. Change Biol. Bioenergy 10, 61–71 (2018).
Article Google Scholar
Maitner, B. S., Boyle, B., Casler, N. & Condit, R. The bien r package: A tool to access the Botanical Information and Ecology Network (BIEN) database. Methods Ecol. Evol. (2018).
ICOS. https://www.icos-cp.eu/ (2022).
Pastorello, G. et al. The FLUXNET2015 dataset and the ONEFlux processing pipeline for eddy covariance data. Sci Data 7, 225 (2020).
Article PubMed PubMed Central Google Scholar
Dorich, C. D. et al. Improving N2O emission estimates with the global N2O database. Curr. Opin. Environ. Sustain. 47, 13–20 (2020).
Article Google Scholar
Lawrence, C. R. et al. An open-source database for the synthesis of soil radiocarbon data: International Soil Radiocarbon Database (ISRaD) version 1.0. Earth Syst. Sci. Data 12, 61–76 (2020).
Article ADS Google Scholar
Schädel, C. et al. Decomposability of soil organic matter over time: the Soil Incubation Database (SIDb, version 1.0) and guidance for incubation procedures. Earth Syst. Sci. Data 12, 1511–1524 (2020).
Article ADS Google Scholar
Jian, J. et al. A restructured and updated global soil respiration database (SRDB-V5). Earth Syst. Sci. Data 13, 255–267 (2021).
Article ADS Google Scholar
Ojima, D., Mosier, A., Del Grosso, S. & Parton, W. J. TRAGNET analysis and synthesis of trace gas fluxes. Global Biogeochem. Cycles 14, 995–997 (2000).
Article ADS CAS Google Scholar
Hibbard, K. A., Law, B. E., Reichstein, M. & Sulzman, J. An analysis of soil respiration across northern hemisphere temperate ecosystems. Biogeochemistry 73, 29–70 (2005).
Article Google Scholar
Horsburgh, J. S. et al. Observations Data Model 2: A community information model for spatially discrete Earth observations. Environ. Model. Soft. 79, 55–74 (2016).
Article Google Scholar
NEON Protocols & Standardized Methods. https://www.neonscience.org/data-collection/protocols-standardized-methods (2020).
EarthChem Data Templates. https://www.earthchem.org/ecl/templates/ (2021).
SLAC. https://www-ssrl.slac.stanford.edu/sfa/ (2022).
WFSFA. https://eesa.lbl.gov/projects/watershed-function-sfa/ (2022).
River Corridor SFA. https://www.pnnl.gov/projects/river-corridor (2022).
AWHSFA. https://www.anl.gov/bio/subsurface-biogeochemical-research (2022).
LLNL Seaborg. https://seaborg.llnl.gov/research/environmental-radiochemistry (2022).
Mercury SFA. https://www.esd.ornl.gov/programs/rsfa/ (2021).
WHONDRS. https://www.pnnl.gov/projects/WHONDRS (2022).
Ameriflux. https://ameriflux.lbl.gov/ (2022).
NEON. https://www.neonscience.org/ (2020).
Vogel, T. M. et al. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 7, 252–252 (2009).
Article CAS Google Scholar
CUAHSI-HIS. Master Controlled Vocabulary Registry for ODM 1.1 http://his.cuahsi.org/mastercvreg/cv11.aspx (2008).
Blodgett, D., Lucido, J. & Kreft, J. Progress on water data integration and distribution: a summary of select US Geological Survey data systems. J. hydroinformatics 18, 226–237 (2016).
Article Google Scholar
NWIS Inventory. https://waterdata.usgs.gov/nwis/inventory (2022).
NEON. https://www.neonscience.org/data-samples/data-management/data-formats-conventions (2020).
WQP User Guide. https://www.waterqualitydata.us/portal_userguide/ (2022).
Garayburu-Caruso, V. A. et al. FTICR, NPOC, TN, and moisture of variably inundated sediment across 48 north American rivers. ESS-DIVE https://doi.org/10.15485/1834208 (2021).
Alves, R. J. E. et al. Kinetic and temperature sensitivity properties of soil exoenzymes through the soil profile down to one-meter depth at a temperate coniferous forest (Blodgett, CA). ESS-DIVE https://doi.org/10.15485/1830417 (2021).
Rogers, A., Ely, K. & Serbin, S. Leaf Photosynthetic Parameters: Quantum Yield, Convexity, Respiration, Gross CO2 Assimilation Rate and Raw Gas Exchange Data, Utqiagvik (Barrow), Alaska, 2016. NGEE Arctic Data Search https://www.osti.gov/biblio/1482338 (2021).
Goldman, A. E., Emani, S. R., Pérez-Angel, L. C., Rodríguez-Ramos, J. A. & Stegen, J. C. Perceived costs and benefits of ICON science and foundational documents associated with “Integrated, Coordinated, Open, and Networked (ICON) science to Advance the Geosciences: Introduction and Synthesis of a Special Collection of Commentary Articles”. (2022).
Roebuck, A. et al. FTICR-MS, Sensor, and Environmental Data from 5 Streams Impacted by the 2020 Holiday Farm Fire Associated with: “Spatiotemporal controls on the delivery of dissolved organic matter to streams following a wildfire”. ESS-DIVE https://doi.org/10.15485/1869708 (2022).
Allison, S. & Martiny, J. B. H. Fungal and bacterial growth variation due to drought and nitrogen addition experimental treatments. Loma Ridge Experimental Project. 2010–2012. ESS-DIVE https://doi.org/10.15485/1828589 (2021).
Dove, N., Torn, M., Hart, S. & Tas, N. Chemistry data from soils and soil incubation experiments from the whole-soil warming experiment at Blodgett Forest, CA, 2018, from: “Metabolic capabilities mute positive response to direct and indirect impacts of warming throughout the soil profile”. ESS-DIVE https://doi.org/10.15485/1866269 (2022).
ESS-DIVE. ESS-DIVE https://ess-dive.lbl.gov/ (2022).
Download references
Robert Crystal-Ornelas, Charuleka Varadharajan, Dylan O’Ryan, Madison Burrus, Shreyas Cholia, Joan Damerow, Valerie C. Hendrix, Zarine Kakalia, Fianna O’Brien, Emily Robles, Maegen Simmonds, Karen Whitenack, and Deborah A. Agarwal were funded through the ESS-DIVE repository by the U.S. DOE’s Office of Science Biological and Environmental Research under contract number DE-AC02-05CH11231. Kim S. Ely and Alistair Rogers were supported through the US Department of Energy contract number DE-SC0012704 to Brookhaven National Laboratory. Michael Crow, Susan Heinz, Terri Velliquette, and Jessica N. Welch were supported through the US Department of Energy contract number DE-AC05-1008 00OR22725 to Oak Ridge National Laboratory. We acknowledge the work of Diana Swantek in producing the Fig. 2 illustration. Reporting format development was supported by through the Office of Biological and Environmental Research in the Department of Energy, Office of Science.
Maegen Simmonds
Present address: Pivot Bio, 2910 Seventh Street, Berkeley, CA, 94710, USA
Robert Crystal-Ornelas
Present address: Github, San Francisco, CA, 94107, USA
Earth and Environmental Sciences Area, Lawrence Berkeley National Laboratory, Berkeley, CA, 94720, USA
Robert Crystal-Ornelas, Charuleka Varadharajan, Dylan O’Ryan, Madison Burrus, Joan Damerow, Zarine Kakalia, Emily Robles, Maegen Simmonds & Karen Whitenack
Environmental Studies Department, California State University, Sacramento, 6000 Jed Smith Dr, Sacramento, CA, 95819, USA
Dylan O’Ryan
Argonne National Laboratory, Lemont, IL, 60439, USA
Kathleen Beilsmith & Pamela Weisenhorn
Pacific Northwest National Laboratory, Joint Global Change Research Institute at the University of Maryland–College Park, College Park, MD, 20740, USA
Benjamin Bond-Lamberty & Stephanie C. Pennington
Environmental Geochemistry Group, SLAC National Accelerator Laboratory, 2575 Sand Hill Road, Menlo Park, CA, 94025, USA
Kristin Boye
Scientific Data Division, Lawrence Berkeley National Laboratory, Berkeley, CA, 94720, USA
Shreyas Cholia, Danielle S. Christianson, Valerie C. Hendrix, Fianna O’Brien & Deborah A. Agarwal
Environmental Sciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, 37830, USA
Michael Crow, Susan L. Heinz, Terri Velliquette & Jessica Nicole Welch
Environmental and Climate Sciences Department, Brookhaven National Laboratory, Upton, NY, 11973, USA
Kim S. Ely & Alistair Rogers
Pacific Northwest National Laboratory, Richland, WA, 99354, USA
Amy E. Goldman
Integrated Life Sciences, Virginia Commonwealth University, Richmond, VA, 23284, USA
Kayla Mathes
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
Conceptualization: D.A.A., C.V., Data curation: R.C.O., C.V., D.O., K.B., B.B.L., K.B., M.C., J.D., K.S.E., A.E.G., S.L.H., K.M., S.C.P., A.R., M.S., T.V., P.W., J.N.W., D.A.A., Funding Acquisition: D.A.A., C.V., Methodology: D.A.A., C.V., R.C.O., J.E.D., K.B., B.B.L., K.B., M.C., J.D., K.S.E., A.E.G., S.L.H., K.M., S.C.P., A.R., M.S., T.V., P.W., J.N.W. Project Administration: D.A.A., K.W. Resources: D.A.A. Software: M.C. Supervision: D.A.A., C.V. Visualization: R.C.O., C.V. Writing – original draft: R.C.O., C.V., J.E.D. Writing – review and editing: R.C.O., C.V., D.O., K.B., B.B.L., K.B., M.B., S.C., D.S.C., M.C., J.D., K.S.E., A.E.G., S.L.H., V.C.H., Z.K., K.M., F.O., S.C.P., E.R., A.R., M.S., T.V., P.W., J.W.N., K.W., D.A.A.
Correspondence to Charuleka Varadharajan.
The authors declare no competing interests.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
Crystal-Ornelas, R., Varadharajan, C., O’Ryan, D. et al. Enabling FAIR data in Earth and environmental science with community-centric (meta)data reporting formats. Sci Data 9, 700 (2022). https://doi.org/10.1038/s41597-022-01606-w
Download citation
Received: 16 May 2022
Accepted: 01 August 2022
Published: 14 November 2022
DOI: https://doi.org/10.1038/s41597-022-01606-w
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Advertisement
© 2022 Springer Nature Limited
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.




